Electron paramagnetic resonance (EPR) can provide structural information on proteins not easily studied by other biophysical techniques such as nuclear magnetic resonance and X-ray crystallography. Inter-residue distances of up to 60Å and residue specific solvent exposure can be measured. Such data can be critical to defining the topology of a protein. However, the amount of information obtained from an EPR dataset is limited by its sparseness and difficulty in relating the EPR measurements into structural properties of the protein.
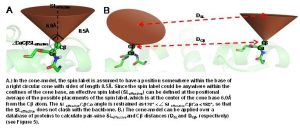
The EPR probe, or spin label, consists of an unpaired electron spin covalently linked at a specified residue position. The spin label projects from the backbone in some unknown orientation so it is nontrivial to relate a distance measured between to spin labels into a distance between two points on the backbone. Additionally, measured solvent accessibility indicates the exposure level of a spin label in relation to solvent. This makes it difficult to derive a relationship between the EPR measurement and the local protein configuration.
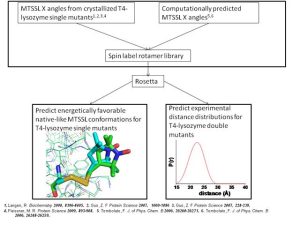
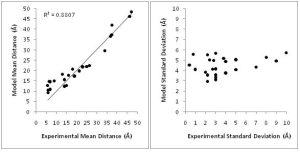
In order to overcome these limitations, we couple EPR data with the BCL protein structure prediction algorithm. The de novo folding aspects of the BCL provide biological information which is not encompassed by the EPR data, while the EPR data improves the efficiency and accuracy of the search for a protein’s structure. A low-resolution cone-model approximates the conformational space available to a spin label and provides the foundation for the BCL to treat the spin label implicitly during protein structure prediction. Having an implicit representation allows the BCL to remain fast and efficient even while leveraging the information content available in EPR data.
Alumni Project Members: Nathan Alexander, Axel Fischer, Steven McIntyre II
