Antibody structural repertoires are determined through diversity of gene segment usage and the introduction of somatic mutations. This structural diversity is responsible for the recognition of an unlimited number of antigens. Traditional techniques for determining structure cannot characterize the breath of structural diversity that these antibody-antigen interactions can adopt because of limited throughput. Using the knowledge-based structural biology program Rosetta, we can use the results from comparative modeling experiments to make high-resolution predictions of protein-protein interactions.
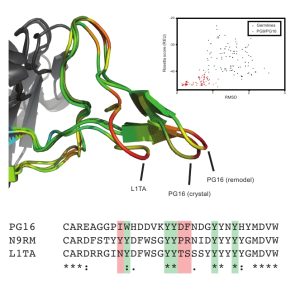
Our focus is on the interactions of antibodies with viral antigens, specifically HIV, flu, and vaccinia. In particular, we hope to generate a broadly neutralizing antibody for the gp120 epitope of the HIV envelope protein; identify epitopes of flu hemagluttinins through homology modeling of antibody/antigen complexes; identify epitopes of antigenic proteins in vaccinia through homology modeling of antibody/antigen complexes; test the viability of the conformation selection hypothesis as applied to antibody maturation. Collaboration with the Crowe Lab at Vanderbilt provides a mechanism for experimental validation of in silico predictions and generation of datasets to train antibody/antigen specific scoring functions.
Energy scoring interactions can help confer the following:
1. Neutralizing potential of a mature antibody.
2. Conserved antigen epitopes on viral membrane proteins.
3. Mutations that need to be made in order to regain neutralization of the antibody.
4. Neutralization threshold mutations during affinity maturation.
5. Rational selections of antibody clones from ultra-high throughput sequencing of an antigen specific antibody repertoire.
Alumni Project Members: Jordan Willis, DavidNannemann, Jens Krause
