A first step towards protein tertiary structure prediction is the identification of secondary structure elements from the sequence. In addition, the identification of trans-membrane spans is required for membrane proteins.
The aim of this project is to simultaneously predict secondary structure and trans-membrane spans with a single tool. The rationale for this approach is the hypothesis that both phenomena are interrelated: When a nascent polypeptide reaches the membrane interface the altered dielectric environment (described by the free energy) leads to the formation of hydrogen bonds and therefore secondary structure. To date there is no single tool available that is suitable for the prediction of both trans-membrane a-helices and ß-strands at the same time.
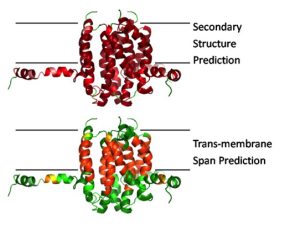
Fig 1:
An Artificial Neural Network (ANN) will be trained on non-redundant datasets of both soluble and membrane proteins (sequence similarity <25%) to accomplish this task (Fig. 1). As input serve several amino acid properties, position-specific scoring matrices from PSIBLAST, and knowledge-based free energies for the secondary structure types helix, strand, or coil and the regions trans-membrane, interface, or solution. A matrix of these numbers over a window of 31 amino acids forms the input for the ANN. The output is a nine-state prediction for the central residue in the sequence window.
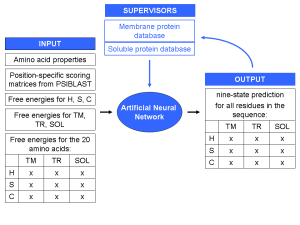
Fig. 2:
Scheme of BCL::Jufo. As amino acid properties the steric parameter, polarizability, volume, iso-electric point, and the solvent-accessible surface area are used as input. The free energies are derived from the membrane protein database.
Alumni Project Members: Julia Koehler Lleman, Ralf Mueller, Nils Woetzel, Mert Karakas
