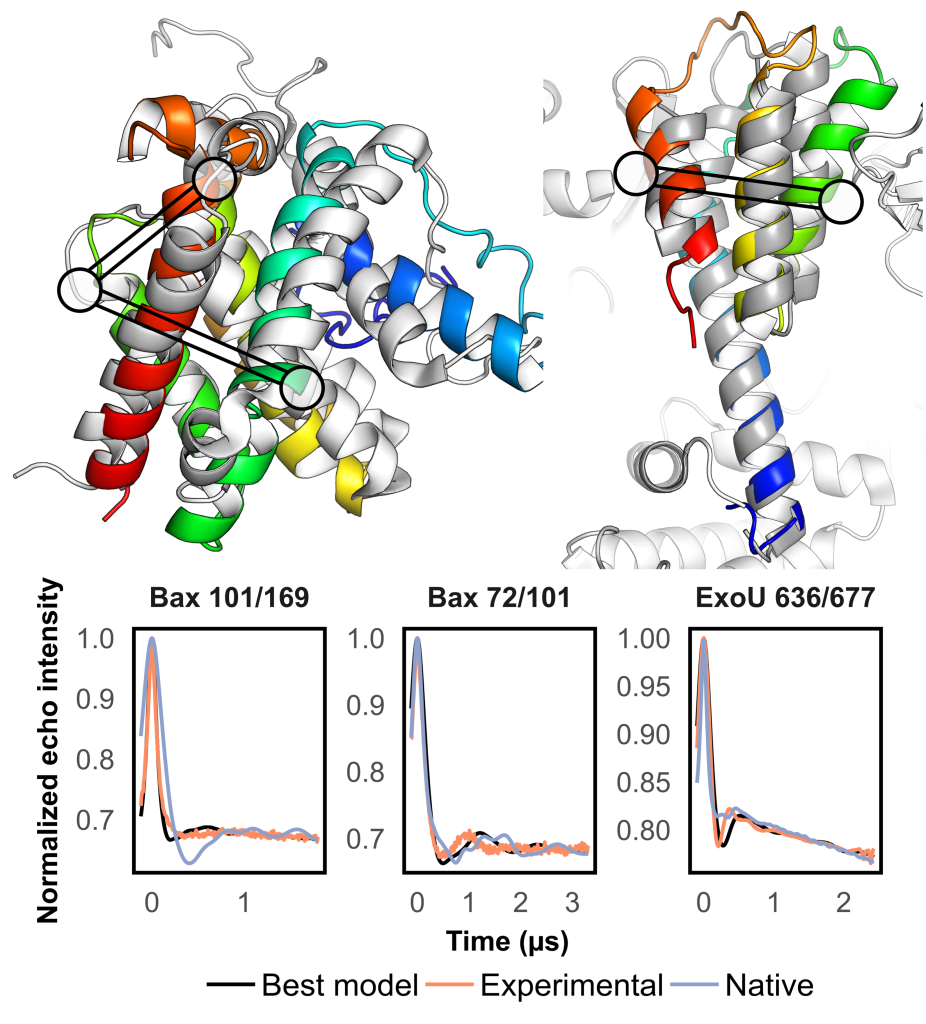
We have an extensive track record of designing methodologies for modeling protein structures using sparse data collected by electron paramagnetic resonance (EPR) spectroscopy. Our work principally focuses on the development of methods for de novo prediction and model refinement. Both RosettaEPR and RosettaDEER allow EPR data to easily be integrated for a variety of protein modeling problems and part of the Rosetta source code. Since their development, we have applied them in collaborations with scientists from around the world to model biomedically relevant proteins such as GPCRs and transporters.
Relevant bibliography:
[1]
Alexander, Nathan, et al. "De novo high-resolution protein structure determination from sparse spin-labeling EPR data." Structure 16.2 (2008): 181-195.
[2]
Del Alamo, Diego, et al. "Rapid simulation of unprocessed DEER decay data for protein fold prediction." Biophysical journal 118.2 (2020): 366-375.
