As a result of all this new structural information, the Personalized Structure Biology is uniquely positioned to identify and triage novel coding variants that appear in clinical settings. As more patients have their genomes sequenced, variants of unknown significance (VUSs) are invariably identified, some of which may be implicated in the patients’ diagnosis. The Personalized Structure Biology can evaluate these VUSs within the context of protein structure, possibly giving insight into the mechanism of disease (e.g. protein destabilization) or of treatment (e.g. disruption of drug target site).
The structural characterization of these VUSs is facilitated through the use of an inhouse, high-throughput analysis pipeline, VUStruct. VUStruct takes as input a list of VUSs (e.g. in the form of genomic coordinates), identify all that are missense coding variants, and then maps them onto all available structural models. VUStruct then applies a set of (user-customizable) analyses to interrogate patient variants using such structure driven methods as:
- Proximity to other known pathogenic variants (PathProx)
- Destabilizing effects on the protein structure
- Evolutionary conservation of amino acids that are proximate in 3D space (COSMIS; github; web app)
As an example of how the structural approach taken by the Personalized Structure Biology can be applied to clinical cases can be seen in a recent diagnosis within the UDN. The proband’s genome harbored a frameshift variant on one copy of fibroblast growth factor binding protein type 2 (FGFBP2). Viewed through the lens of structural biology, this VUS was predicted to ablate secondary structure in the protein (see Figure 1). This structural consequence would be predicted to inhibit the ability of FGFBP2 to bind fibroblast growth factor, thus providing a mechanistic model of pathogenicity.
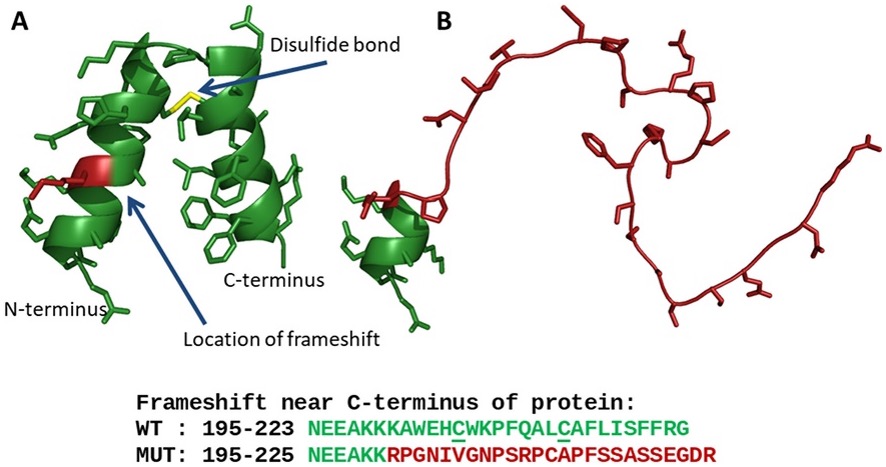
Figure 1. A structural model for FGFBP2. The model for FGFBP2 suggests a mechanism for the effect of the frameshift variant present in the proband. (A) The C-terminal domain is predicted to form two alpha helices held at an angle by a conserved disulfide bond. Protein backbone is shown as green ribbons; sidechains are shown as sticks; position of frameshift is highlighted in red. (B) The frameshifted sequence is predicted to form a disordered random coil. (from: Newman et al. Mol Genet Genomic Med. 2019)
Select Personalized Structure Biology Publications:
[1]
Shibao CA, Joos K, Phillips JA 3rd, Cogan J, Newman JH, Hamid R, Meiler J, Capra J, Sheehan J, Vetrini F, Yang Y, Black B, Diedrich A, Roberston D, Biaggioni I. Familial Autonomic Ganglionopathy Caused by Rare CHRNA3 Genetic Variants. Neurology. 2021 Jul 13;97(2):e145-e155.
[2]
Newman JH, Shaver A, Sheehan JH, Mallal S, Stone JH, Pillai S, Bastarache L, Riebau D, Allard-Chamard H, Stone WM, Perugino C, Pilkinton M, Smith SA, McDonnell WJ, Capra JA, Meiler J, Cogan J, Xing K, Mahajan VS, Mattoo H, Hamid R, Phillips JA 3rd; Undiagnosed Disease Network. IgG4-related disease: Association with a rare gene variant expressed in cytotoxic T cells. Mol Genet Genomic Med. 2019 Jun;7(6):e686. doi: 10.
[3]
Sivley RM, Sheehan JH, Kropski JA, Cogan J, Blackwell TS, Phillips JA, Bush WS, Meiler J, Capra JA. Three-dimensional spatial analysis of missense variants in RTEL1 identifies pathogenic variants in patients with Familial Interstitial Pneumonia. BMC Bioinformatics. 2018 Jan 23;19(1):18
