Fragment Assembly of RNA with Full Atom Refinement (FARFAR) is a framework within the Rosetta suite that can produce de novo or homologous RNA structure models and has been improved with the release of FARFAR2, enabling larger target RNAs to be modeled [1],[2]. The target RNA sequence and predicted secondary structure or known homologies are utilized as inputs prior to fragment assembly. Constructs that pass quality control filters are then subjected to full atom refinement prior to producing clustered ensembles. Additional information on FARFAR2 can be found at:
https://www.rosettacommons.org/docs/latest/FARFAR2
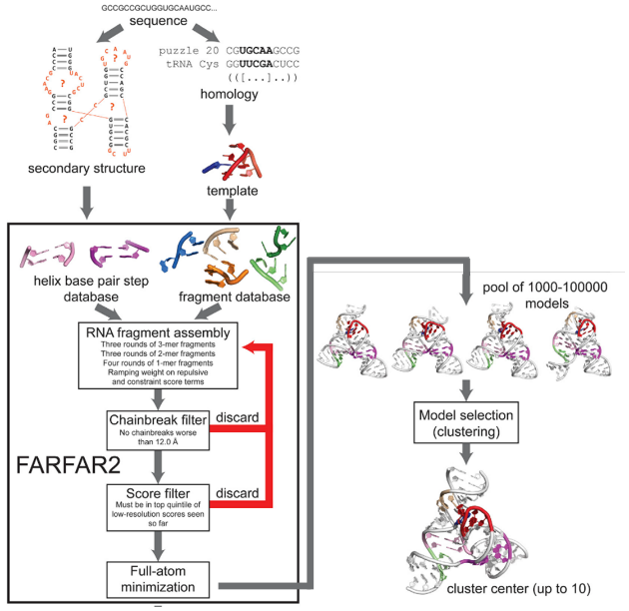
Figure 1: Overview of FARFAR2 Modeling Workflow. Shown here is each step required for modeling RNA using Rosetta FARFAR2, starting from RNA sequence. In addition to as mentioned previously, the secondary structures can be generated utilizing experimental biochemical data to improve predictions prior to FARFAR2 modeling. Image modified from Figure 1 Watkins et al., 2020
Relevant Bibliography:
[1]
Das, R., Karanicolas, J., and Baker, D. (2010), "Atomic accuracy in predicting and designing noncanonical RNA structure". Nature Methods 7:291-294.
[2]
Watkins, A. M.; Rangan, R.; Das, R. “FARFAR2: Improved de novo Rosetta prediction of complex global RNA folds.” Structure, 2020, 28: 963-976.
