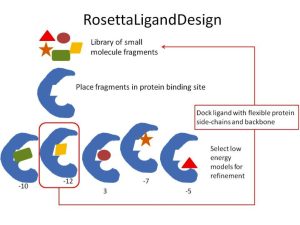
Small molecule design consists of assembly, docking, and scoring. Since Rosetta already has a docking algorithm (RosettaLigand) and a scoring framework, all that is needed for small molecule design is an assembly algorithm. As an initial approach, we have chosen random assembly of fragments from an input library. The algorithm is represented in figure 1. Each cycle of design consists of extending the small molecule with random fragments chosen from a library, filtering small molecule designs based on energy score, and docking the new ligand using RosettaLigand.
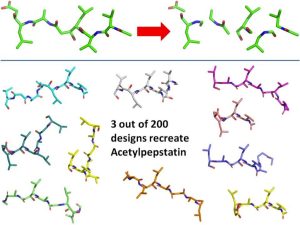
In figure two we test our Rosetta assembly code. In the top panel acetylpepstatin was split into 6 fragments Five cycles of random assembly were used to generate the small molecules seen in the bottom panel.
Current Project Members: Rocco Moretti
Alumni Project Members: Gordon Lemmon, Dan Viox, Kristin Glab, Steven Combs
