The complementary-determining regions (CDR) loops in heavy and light chain of antibodies play an important role in antigen recognition. However, the high variability in the CDR loop regions has posed a significant challenge in accurate structure prediction and design of antibodies. We use the RosettaAntibody application (<>), AbPredict and RosettaCM protocol within the Rosetta Suite to perform antibody loop modeling. Additionally, we have developed a method of computational design of cyclic peptides based on the structure of an antibody hypervariable loop to generate mini-antibodies. We hypothesized that by designing peptides to mimic antibody loops, we could enhance its breadth of binding by reducing contact with hypervariable antigenic residues, and improve binding affinity and specificity by incorporating non-canonical amino acids. [[ADD LINKS]]
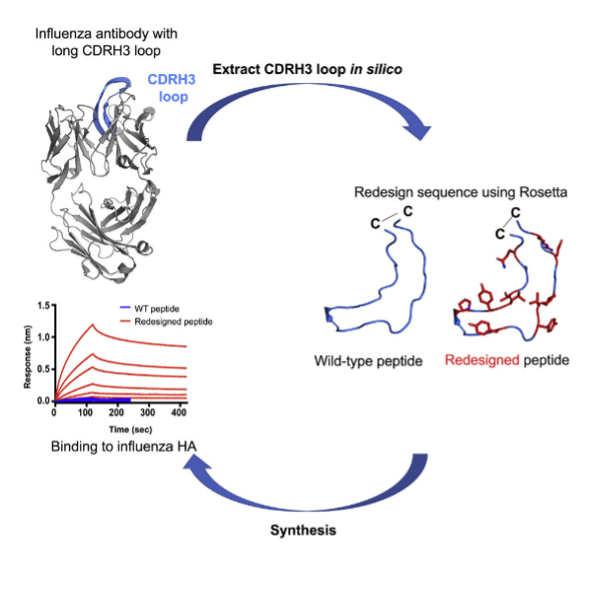
Figure 1: Design of cyclic peptides based on the structure of an antibody hypervariable loop
Bibliography
[1]
Weitzner BD, Gray JJ. Accurate Structure Prediction of CDR H3 Loops Enabled by a Novel Structure-Based C-Terminal Constraint. J Immunol. 2017;198(1):505-515. doi:10.4049/jimmunol.1601137
[2]
Sevy AM, Gilchuk IM, Brown BP, Bozhanova NG, Nargi R, Jensen M, Meiler J, Crowe JE Jr. Computationally Designed Cyclic Peptides Derived from an Antibody Loop Increase Breadth of Binding for Influenza Variants. Structure. 2020 Oct 6;28(10):1114-1123.e4. doi: 10.1016/j.str.2020.04.005.
